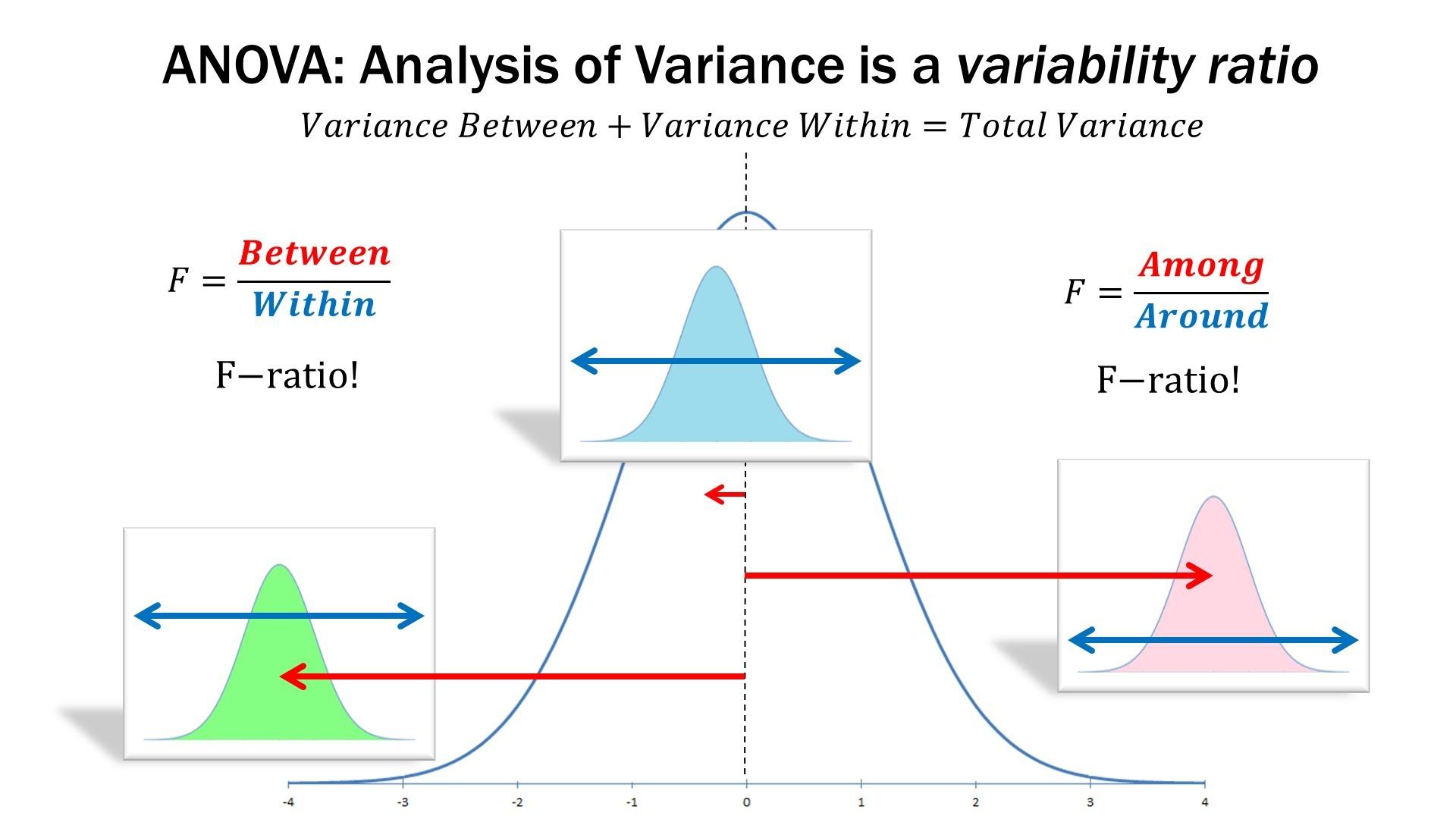
估计阅读时长: 14 分钟一般而言,如果我们在进行组学数据分析的时候,如果想要比较两组数据之间是否存在有差异性,一般是对两两比较的两组数据进行T-检验。但是在代谢组学数据分析领域内,则很多的组学数据分析情况为比较两组以上的数据,寻找差异的biomarker。那这个时候就需要使用上ANOVA统计检验方法了。 Order by Date Name Attachments anova • 105 kB • 844 click 2022年5月28日ANOVA-screen • 27 […]
Recent Posts
Archives
- February 2026 (6)
- January 2026 (2)
- December 2025 (10)
- November 2025 (2)
- October 2025 (1)
- August 2025 (3)
- July 2025 (2)
- June 2025 (6)
- May 2025 (3)
- November 2023 (1)
- June 2023 (2)
- May 2023 (2)
- April 2023 (2)
- March 2023 (2)
- February 2023 (1)
- August 2022 (2)
- July 2022 (2)
- June 2022 (5)
- May 2022 (5)
- April 2022 (4)
- March 2022 (3)
- January 2022 (2)
- December 2021 (2)
- November 2021 (2)
- October 2021 (6)
- September 2021 (8)
- August 2021 (8)
- July 2021 (6)
- June 2021 (20)
- May 2021 (10)
Tags
algorithm (33)
bilibili (3)
binary tree (4)
clustering (21)
contour (3)
Darwinism (4)
dataframe (3)
data visualization (23)
dotnet-core (25)
GCModeller (20)
gdi+ (23)
gem (7)
ggplot (14)
graph (14)
heatmap (5)
http (4)
image processing (7)
kegg (8)
kmeans (3)
language (7)
linq (3)
linux (8)
machine learning (4)
mass spectrometry (12)
math (19)
metagenomics (6)
motif (4)
MSI (4)
mzkit (19)
network (8)
pathway (4)
pipeline (4)
query (5)
R# (44)
rsharp (23)
scripting (14)
single-cell (6)
sql (3)
symbolic computation (3)
text processing (4)
typescript (3)
ubuntu (4)
uniprot (3)
vb (19)
VisualBasic (50)


[…] 在前面的一篇《基因组功能注释(EC Number)的向量化嵌入》博客文章中,针对所注释得到的微生物基因组代谢信息,进行基于TF-IDF的向量化嵌入之后。为了可视化向量化嵌入的效果,通过UMAP进行降维,然后基于降维的结果进行散点图可视化。通过散点图可视化可以发现向量化的嵌入结果可以比较好的将不同物种分类来源的微生物基因组区分开来。 […]
😲啊?
谢老师,写快点呀,在看着你更新文章呢。
[…] 最近的工作中我需要按照之前的这篇博客文章《基因组功能注释(EC Number)的向量化嵌入》中所描述的流程,将好几十万个微生物基因组的功能蛋白进行酶编号的比对注释,然后基于注释结果进行向量化嵌入然后进行数据可视化。通过R#脚本对这些微生物基因组的蛋白fasta序列的提取操作,最终得到了一个大约是58GB的蛋白序列。然后将这个比较大型的蛋白序列比对到自己所收集到的ec number注释的蛋白序列参考数据库之上。 […]
[…] […]