文章阅读目录大纲
 https://github.com/rsharp-lang/ggplot
https://github.com/rsharp-lang/ggplot
接上一篇博客文章中谈到,我们已经通过R#语言之中的ggplot程序包绘制出了一个可以使用的火山图。在这里,我们将会通过在火山图上添加更多的可视化元素来为大家介绍R#语言之中的ggplot程序包的进阶使用方式。
为数据点添加文本标签
为图表上的数据点添加文本标签,非常的容易,我们添加了一个geom_text或者geom_label图层即可。
ggplot(volcano, aes(x = "log2FC", y = "p.value"), padding = "padding:250px 500px 250px 300px;")
+ geom_point(aes(color = "factor"), color = "black", shape = "circle", size = 25)
+ scale_colour_manual(values = list(
Up = "red",
"Not Sig" = "gray",
Down = "steelblue"
))
+ geom_text(aes(label = "ID"))
+ labs(x = "log2(FoldChange)", y = "-log10(P-value)")
+ ggtitle("Volcano Plot (A vs B)")
+ scale_x_continuous(labels = "F2")
+ scale_y_continuous(labels = "F2")
;上面的这一段代码是百分之百可行的,但是样式嘛,大家可以自行体会:

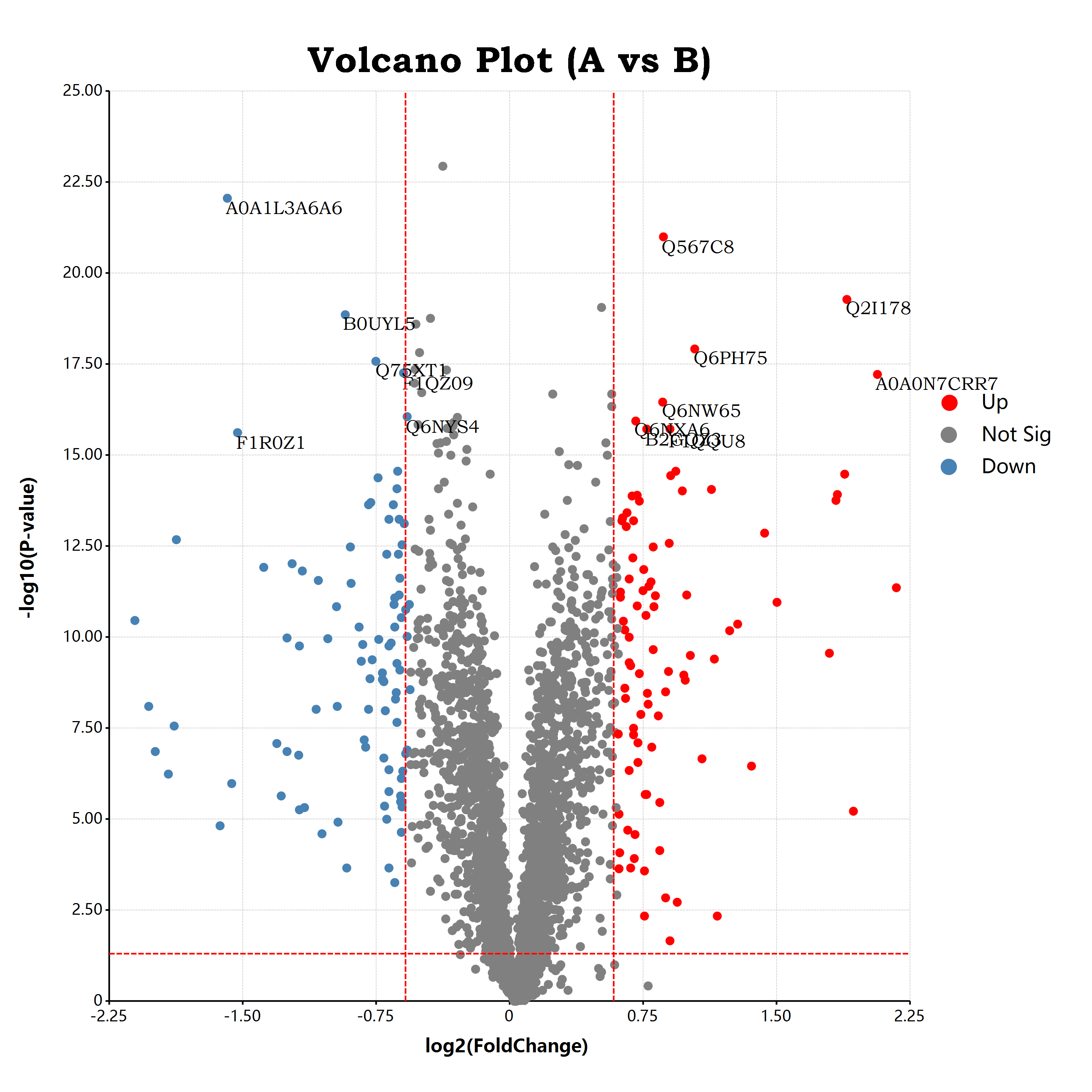
呃呃呃呃,因为我们默认是将所有的标签都显示出来的,所以图上面的标签数量有点多。那,这个时候就会需要使用到ggplot之中的进阶功能了:筛选器
对文本标签使用筛选器
那现在我们需要将所有差异显著的基因的编号都显示出来。这个要怎么做呢?在R语言之中的ggplot程序包,大家进行选择性的绘制标签一般会需要在dataframe之中提前做好筛选操作,例如:
# [1] "peeks of the raw data:"
# ID p.value log2FC factor
# <mode> <string> <double> <double> <string>
# [1, ] "Q5XJ10" 9.81 -0.205 "Not Sig"
# [2, ] "A8WG05" 5.21 0.0472 "Not Sig"
# [3, ] "Q8JH71" 11.6 -0.38 "Not Sig"
# [4, ] "Q7T3L3" 8.85 0.165 "Not Sig"
# [5, ] "Q567C8" 21 0.837 "Up"
# [6, ] "Q92005" 0.091 -0.00514 "Not Sig"
volcano[, "labels"] = ifelse((volcano[, "factor"] != "Not Sig") && (volcano[, "p.value"] >= 15), volcano[, "ID"], NULL);但是在R#语言的ggplot程序包之中,大家可以更加简单的应用筛选器功能来选择所需要绘制的标签文本了:
geom_text(aes(label = "ID"), which = ~(factor != "Not Sig") && (p.value >= 15) )我们可以直接通过which参数对ID标签文本进行筛选:将所有的factor不为Not Sig,以及pvalue极显著的文本标签显示出来:

ggplot对筛选器的底层代码实现
在ggplot程序包中的图层对象,都具有一个which筛选器表达式对象。这样子我们在进行图层绘制的时候,就可以对每一个图层应用自己单独的筛选器了。
Public MustInherit Class ggplotLayer
Public Property which As Expression
Public Function getFilter(ggplot As ggplot) As BooleanVector
Dim i As New List(Of Object)
Dim measure As New Environment(ggplot.environment, ggplot.environment.stackFrame, isInherits:=False)
Dim x = DirectCast(ggplot.data, dataframe).colnames _
.SeqIterator _
.ToArray
For Each var In x
Call measure.Push(var.value, Nothing, [readonly]:=False)
Next
For Each row As NamedCollection(Of Object) In DirectCast(ggplot.data, dataframe).forEachRow(x.Select(Function(xi) xi.value).ToArray)
For Each var In x
Call measure(var.value).SetValue(row(var), measure)
Next
i.Add(REnv.single(which.Evaluate(measure)))
Next
Return New BooleanVector(REnv.asLogical(i.ToArray))
End Function
End Class在上面我们对文本标签所应用的筛选器表达式(factor != "Not Sig") && (p.value >= 15)
(factor != "Not Sig") && (p.value >= 15)
# is equals to vector expression
volcano[, "labels"] = ifelse((volcano[, "factor"] != "Not Sig") && (volcano[, "p.value"] >= 15), volcano[, "ID"], NULL);
# [1] "peeks of the raw data:"
# ID p.value log2FC factor
# <mode> <string> <double> <double> <string>
# [1, ] "Q5XJ10" 9.81 -0.205 "Not Sig"
# [2, ] "A8WG05" 5.21 0.0472 "Not Sig"
# [3, ] "Q8JH71" 11.6 -0.38 "Not Sig"
# [4, ] "Q7T3L3" 8.85 0.165 "Not Sig"
# [5, ] "Q567C8" 21 0.837 "Up"
# [6, ] "Q92005" 0.091 -0.00514 "Not Sig"上面的两个表达式所要实现的功能是一样的,但是很明显ggplot程序包之中的筛选器表达式更加的精炼。那如何实现上面的表达式求值呢。在ggplot的底层代码之中,每一个图层对象都有一个各自的getFilter
Dim measure As New Environment(ggplot.environment, ggplot.environment.stackFrame, isInherits:=False)
Dim x = DirectCast(ggplot.data, dataframe).colnames _
.SeqIterator _
.ToArray
For Each var In x
Call measure.Push(var.value, Nothing, [readonly]:=False)
Next那有了这些符号之后呢,我们接下来就只需要对原始数据框中的每一行数据都代入到计算环境之中做筛选器表达式求值即可:
For Each row As NamedCollection(Of Object) In DirectCast(ggplot.data, dataframe).forEachRow(x.Select(Function(xi) xi.value).ToArray)
For Each var In x
Call measure(var.value).SetValue(row(var), measure)
Next
i.Add(REnv.single(which.Evaluate(measure)))
Next最后呢,我们只需要使用所得到的筛选标记对原始数据做筛选即可:
If Not which Is Nothing Then
Dim i As BooleanVector = getFilter(ggplot)
labelList = (New Vector(Of Label)(labelList))(i)
anchors = (New Vector(Of Anchor)(anchors))(i)
End If在图表中添加分界线
接下来,在完成了标签的添加之后,我们还需要为差异表达的基因以及非差异表达基因的散点之间添加分界线。分界线的添加,我们可以直接使用abline图层来完成。可以分别通过geom_hline添加横线,geom_vline添加竖线。所谓的abline,就是由a和b两个点所组成的一条直线线段。下面所示的代码用于分别添加log2FC以及p-value的分界线:
geom_hline(yintercept = -log10(0.05), color = "red", line.width = 5, linetype = "dash")
+ geom_vline(xintercept = log2(foldchange), color = "red", line.width = 5, linetype = "dash")
+ geom_vline(xintercept = -log2(foldchange), color = "red", line.width = 5, linetype = "dash")分界线的底层实现代码
以横线图层为例,调用geom_hline函数实际上就是创建一个abline图层:
<ExportAPI("geom_hline")>
Public Function geom_hline(yintercept As Double,
Optional color As Object = "black",
Optional line_width! = 2,
Optional linetype As DashStyle = DashStyle.Solid) As ggplotLayer
Dim a As New PointF(Single.MinValue, yintercept)
Dim b As New PointF(Single.MaxValue, yintercept)
Dim style As New Pen(RColorPalette.getColor(color).TranslateColor, line_width) With {
.DashStyle = linetype
}
Return New ggplotABLine With {
.abline = New Line(a, b, style)
}
End Function之后在进行绘制出来即可:
Public Class ggplotABLine : Inherits ggplotLayer
Public Property abline As Line
Public Overrides Function Plot(g As IGraphics,
canvas As GraphicsRegion,
baseData As ggplotData,
x() As Double,
y() As Double,
scale As DataScaler,
ggplot As ggplot,
theme As Theme) As legendGroupElement
Dim a As PointF = constraint(abline.A, scale)
Dim b As PointF = constraint(abline.B, scale)
a = scale.Translate(a)
b = scale.Translate(b)
Call g.DrawLine(abline.Stroke, a, b)
Return Nothing
End Function
Private Shared Function constraint(pf As PointF, scale As DataScaler) As PointF
Dim x As Single = If(pf.X < scale.xmin, scale.xmin, pf.X)
Dim y As Single = If(pf.Y < scale.ymin, scale.ymin, pf.Y)
Return New PointF With {
.X = If(x > scale.xmax, scale.xmax, x),
.Y = If(y > scale.ymax, scale.ymax, y)
}
End Function
End Class
完整的绘图脚本
下面是在这篇博客文章中所展示的筛选器功能的完整的R#脚本代码:
require(ggplot);
const volcano = read.csv(`${@dir}/log2FC.csv`);
const foldchange = 1.5;
# create color factor for scatter points
volcano[, "factor"] = ifelse(volcano[, "log2FC"] > log2(foldchange), "Up", "Not Sig");
volcano[, "factor"] = ifelse(volcano[, "log2FC"] < -log2(foldchange), "Down", volcano[, "factor"]);
volcano[, "factor"] = ifelse(volcano[, "p.value"] < 0.05, volcano[, "factor"], "Not Sig");
# transform of the pvalue scale
volcano[, "p.value"] = -log10(volcano[, "p.value"]);
print("peeks of the raw data:");
print(head(volcano));
print("count of the factors:");
print(`Up: ${sum("Up" == volcano[, "factor"])}`);
print(`Not Sig: ${sum("Not Sig" == volcano[, "factor"])}`);
print(`Down: ${sum("Down" == volcano[, "factor"])}`);
# [1] "peeks of the raw data:"
# ID p.value log2FC factor
# <mode> <string> <double> <double> <string>
# [1, ] "Q5XJ10" 9.81 -0.205 "Not Sig"
# [2, ] "A8WG05" 5.21 0.0472 "Not Sig"
# [3, ] "Q8JH71" 11.6 -0.38 "Not Sig"
# [4, ] "Q7T3L3" 8.85 0.165 "Not Sig"
# [5, ] "Q567C8" 21 0.837 "Up"
# [6, ] "Q92005" 0.091 -0.00514 "Not Sig"
#
# [1] "count of the factors:"
# [1] "Up: 90"
# [1] "Not Sig: 2643"
# [1] "Down: 93"
bitmap(file = `${@dir}/volcano.png`, size = [3000, 3000]) {
# create ggplot layers and tweaks via ggplot style options
ggplot(volcano, aes(x = "log2FC", y = "p.value"), padding = "padding:250px 500px 250px 300px;")
+ geom_point(aes(color = "factor"), color = "black", shape = "circle", size = 25)
+ scale_colour_manual(values = list(
Up = "red",
"Not Sig" = "gray",
Down = "steelblue"
))
+ geom_text(aes(label = "ID"), which = ~(factor != "Not Sig") && (p.value >= 15) )
+ geom_hline(yintercept = -log10(0.05), color = "red", line.width = 5, linetype = "dash")
+ geom_vline(xintercept = log2(foldchange), color = "red", line.width = 5, linetype = "dash")
+ geom_vline(xintercept = -log2(foldchange), color = "red", line.width = 5, linetype = "dash")
+ labs(x = "log2(FoldChange)", y = "-log10(P-value)")
+ ggtitle("Volcano Plot (A vs B)")
+ scale_x_continuous(labels = "F2")
+ scale_y_continuous(labels = "F2")
;
}- 单细胞视角下的微生物基因组代谢酶嵌入分析 - 2026年2月25日
- 基因组代谢酶层级嵌入 - 2026年2月23日
- 基因组功能吉布斯LDA主题建模 - 2026年2月23日


One response
[…] 《【ggplot】在R#语言之中的进阶火山图》 […]